Ulti-HEP: A Breakthrough Alternative to Primary and Immortalized Liver Cells
Redefining iPSC-Derived Liver Models for Drug Discovery and Disease Research
Meeting an Urgent Industry Need
Drug development is increasingly hindered by a lack of predictive, scalable, and functionally accurate liver models. Traditional in vitro systems such as primary human hepatocytes, suffer from poor longevity, inconsistency, and donor variation leading to low reproducibility. Meanwhile, immortalised cancer cell lines do not offer a phenotypically relevant enough model for efficacy investigations. While iPSC (induced pluripotent stem cell) technology holds promise, conventional iPSC-derived hepatocytes often fall short, displaying immature phenotypes and limited metabolic activity.
DefiniGEN’s next-generation platform directly overcomes these limitations—offering a new standard in hepatocyte functionality and reproducibility.

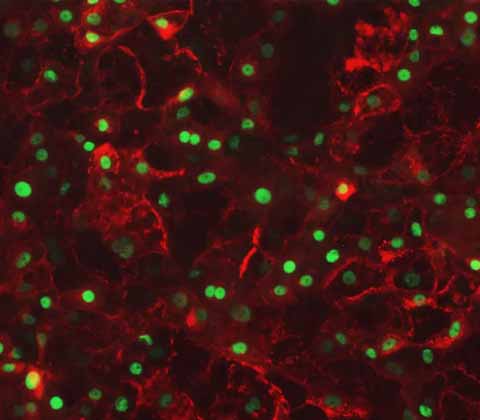
The Ulti-HEP Advantage:
Built upon the pioneering work of Prof Ludovic Vallier, DefiniGEN’s platform represents a significant evolution in iPSC-derived hepatocyte production. Our proprietary 3-step iPSC differentiation protocol has been extensively refined, yielding Ulti-HEP, a best-in-class iPSC-derived hepatocyte with:
-
Enhanced functional maturity
-
Sourced from a single donor
-
Consistent, scalable batch production
-
High-fidelity gene and protein expression profiles
This innovation empowers researchers to make more confident, translationally relevant decisions at the start of their drug discovery and development pipeline.
Applications Across the R&D Pipeline
Versatile, Predictive, and Scalable iPSC-Derived Hepatocytes for Every Stage of Development
DefiniGEN’s Ulti-HEP platform is purpose-built to meet the evolving demands of modern drug discovery, toxicology, and disease research. Whether you're exploring disease mechanisms, screening compound libraries, or developing patient-specific models, our wild-type and disease models deliver the physiological relevance and consistency needed to improve translational outcomes and reduce reliance on animal models.
Disease Modeling
Accurately replicate human liver diseases using patient-derived iPSCs to explore pathophysiology and therapeutic interventions.
Drug Safety & Toxicology
Screen compounds for hepatotoxicity and off-target effects using functionally predictive liver cells.
Mechanism of Action Studies
Uncover drug targets, validate biological pathways, and understand compound effects at a cellular level.
Personalized medicine
Predict individual patient responses through tailored hepatocyte models derived from donor-specific iPSCs.
Ulti-HEP is demonstrably superior to conventional iPSC-derived hepatocytes
Through Principal Component Analysis (PCA) we demonstrate that Ulti-HEP clusters closer to Primary human hepatocytes when compared to conventional iPSC-hepatocytes.
Conventional vs. DefiniGEN iPSC-derived hepatocytes



Conventional vs. DefiniGEN iPSC-derived hepatocytes:
Comparing metabolic pathways



Frequently asked questions
Do Ulti-HEPs express ASGR1?
Yes, ASGR1 and ASGR2 has been detected by qPCR analysis and ICC and has been shown to localize on the cell membrane.
Do they have a functional urea cycle?
Yes this has been characterized by gene expression profiling, Western blot analysis and Ornithine stimulated functional analysis.
Can you see CYP expression activity?
Expression of CYPs is detectable by qPCR analysis, but they are lower than Primary Human Hepatocytes (PHH).

Let’s work together.
Contact us today to speak to one of our experts about how Ulti-HEP can accelerate your research

